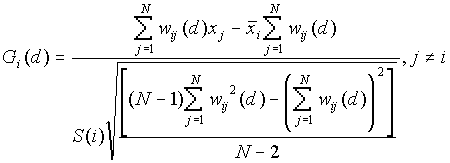 [1]
[1]Local Gi(d) and Gi*(d)
Gi(d) and Gi*(d) are described by Ord and Getis (1995). They indicate the extent to which a location is surrounded by a cluster of high or low values. The Gi(d) statistic excludes the value at i from the summation and is used for spread or diffusion studies, while the Gi*(d) includes the value at i in the summation and is most often used for studies of clustering.
Input
You’ll be asked to enter the input data file. This file should contain N rows coordinates, and the corresponding value of the test variable (x).
Analysis
The null hypothesis in both of these tests is that there is no association between the value found at one site and its neighbors within the specified distance. The expected value under the null hypothesis is 0, and the variance is 1. Therefore, the Gi(d) or Gi*(d ) statistics may be examined as a standard normal variate. Positive Gi(d) or Gi*(d) indicate spatial association of high values, whereas negative Gi(d) or Gi*(d) indicate spatial association of low values.
Formula
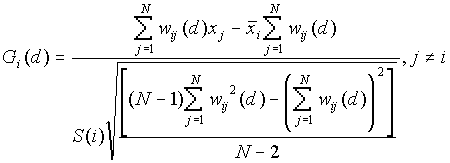 [1]
[1]
where
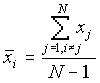
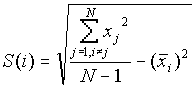
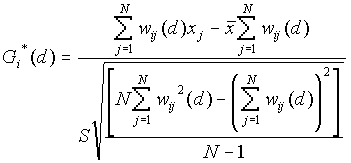 [2]
[2]
where
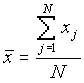
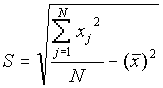
Output
The output file includes the total number of points in the analysis, and a listing of the Gi(d) or Gi*(d) value for each point at each distance specified.
Example Gi*(d)
For this example we will examine the clustering of high versus low cancer rates in New Mexico’s counties using the Gi*(d) statistic. The data are taken from the National Cancer Institute Biometry Research Group Datasets, and it is available on their website (
http://dcp.nci.nih.gov/bb/datasets.html). The total number of cases diagnosed between 1980 and 1989 were divided by the 1985 population estimate to determine the cancer rate (cases per 10,000 population) of each county. These data are shown in Table 1. The points used for this analysis are the county seats, and the coordinate units are kilometers. Figure 1 is a map of these county seatsTable 1: Cancer Rates of New Mexico Counties
|
ID Number |
County |
Coordinates |
Cancer Rate |
||
|
1 |
Bernalillo |
219 |
338 |
40.29 |
|
|
2 |
Catron |
027 |
189 |
33.12 |
|
|
3 |
Chaves |
418 |
156 |
76.91 |
|
|
4 |
Colfax |
408 |
538 |
47.37 |
|
|
5 |
Curry |
534 |
262 |
35.19 |
|
|
6 |
DeBaca |
438 |
272 |
73.98 |
|
|
7 |
DonaAna |
212 |
037 |
36.67 |
|
|
8 |
Eddy |
451 |
037 |
68.35 |
|
|
9 |
Grant |
073 |
083 |
37.90 |
|
|
10 |
Guadelupe |
428 |
322 |
31.24 |
|
|
11 |
Harding |
458 |
418 |
27.23 |
|
|
12 |
Hidalgo |
033 |
040 |
44.43 |
|
|
13 |
Lea |
528 |
103 |
41.38 |
|
|
14 |
Lincoln |
295 |
179 |
63.39 |
|
|
15 |
Los Alamos |
246 |
425 |
25.56 |
|
|
16 |
Luna |
119 |
030 |
80.02 |
|
|
17 |
McKinley |
030 |
385 |
11.78 |
|
|
18 |
Mora |
335 |
435 |
42.69 |
|
|
19 |
Otero |
289 |
100 |
32.19 |
|
|
20 |
Quay |
481 |
345 |
68.23 |
|
|
21 |
Rio Arriba |
222 |
514 |
24.21 |
|
|
22 |
Roosevelt |
518 |
239 |
38.90 |
|
|
23 |
Sandoval |
229 |
365 |
48.29 |
|
|
24 |
San Juan |
096 |
531 |
27.30 |
|
|
25 |
San Miguel |
345 |
398 |
31.54 |
|
|
26 |
SantaFe |
279 |
402 |
34.77 |
|
|
27 |
Sierra |
166 |
127 |
140.73 |
|
|
28 |
Socorro |
199 |
222 |
37.79 |
|
|
29 |
Taos |
315 |
484 |
26.60 |
|
|
30 |
Torrance |
272 |
302 |
66.59 |
|
|
31 |
Union |
521 |
488 |
52.70 |
|
|
32 |
Valencia |
160 |
329 |
45.19 |
|
Figure 1: New Mexico County Seats 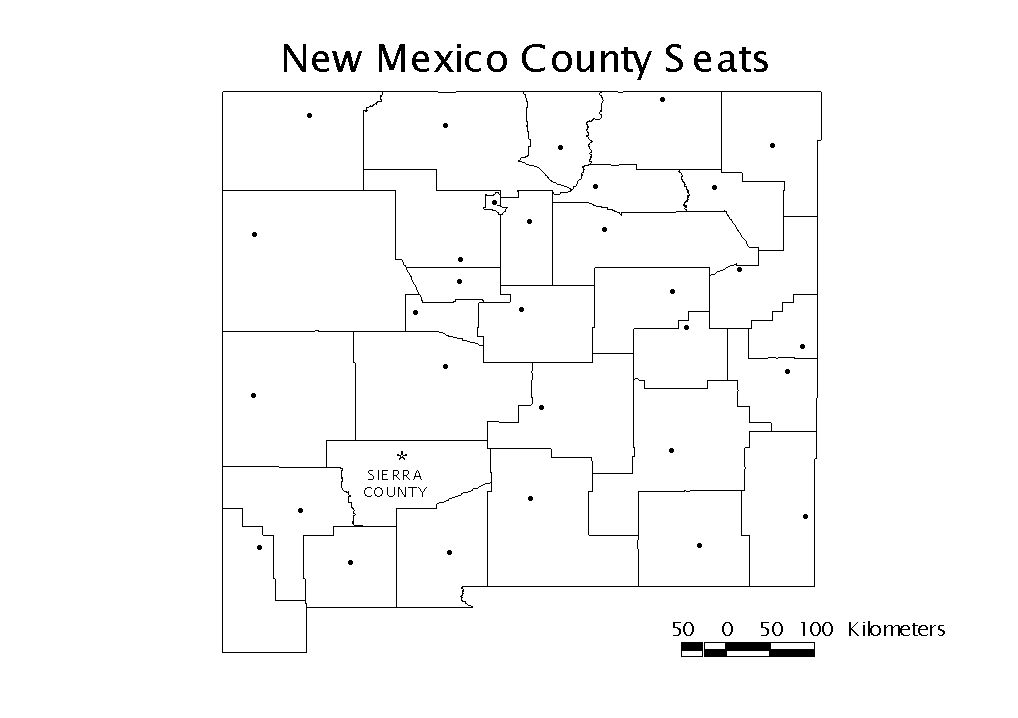
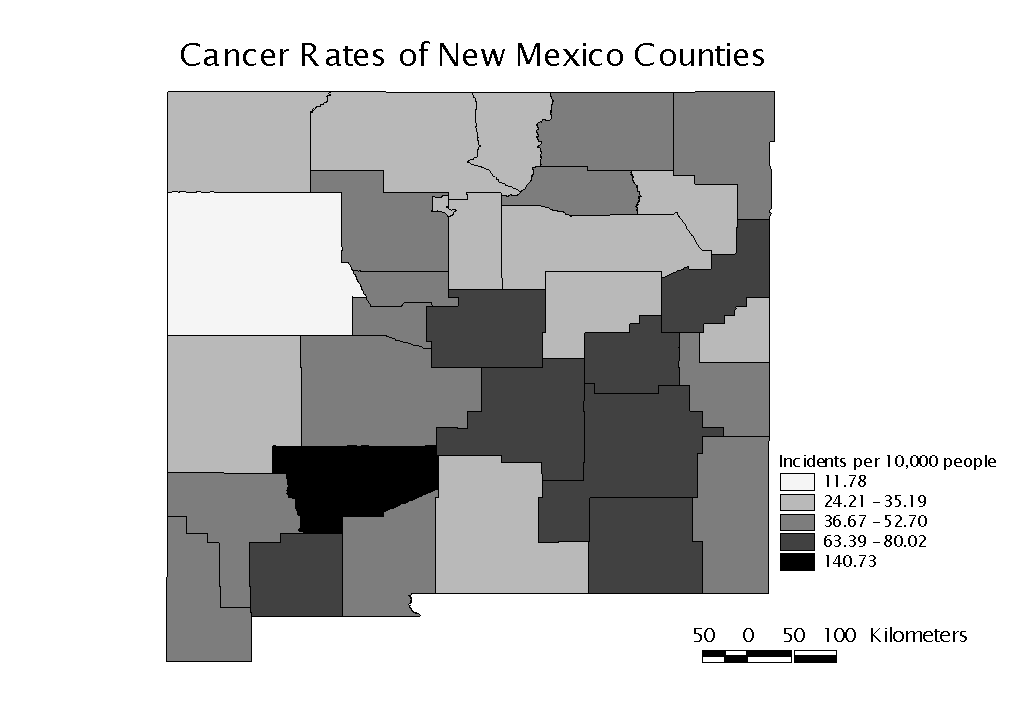
Sierra County, with a rate of 140.73 per 10,000 population, has a much higher rate than all other counties. The next highest cancer rate is 80.02 cases per 10,000 people in Luna County. We will examine the county data using the G* statistic to determine if this disparity stands alone or is the center of a cluster. Figure 2 is a map of the distribution of cancer rates by county.
Figure 2: Cancer rates of New Mexico Counties
A summary table of the Gi*(d) values for Sierra County is shown in Table 2. Recall that the Gi*(d) statistic is based on accumulated clustering tendencies. The complete output file is included in Table 3.
Table 2: Gi* values for Sierra County (observation #27)
|
Distance(d) |
50km |
100km |
150km |
200km |
250km |
300km |
|
Gi*(d) |
3.95 |
3.95 |
1.81 |
1.40 |
1.49 |
1.54 |
Clustering is in evidence at 50km and 100km, but as distance increases the tendancy for clustering decreases. Since the next closest point to the Sierra County Seat is over 100km away, this indicates that Sierra County stands alone as a cluster. As the neighboring counties are considered the Gi*(d) values drop, and this implies that anti-clustering forces are in command.
Table 3: Output File
The input data file: nmlung.txt
The total number of points: 32
Distance: 50.000000
# Gi*(d)
1 -0.14201
2 -0.56799
3 1.27146
4 0.03060
5 -0.57951
6 1.14838
7 -0.41887
8 0.91189
9 -0.36720
10 -0.64696
11 -0.81540
12 -0.09290
13 -0.22102
14 0.70354
15 -0.99498
16 1.40210
17 -1.46440
18 -0.57529
19 -0.60705
20 0.90685
21 -0.94226
22 -0.57951
23 -0.14201
24 -0.81246
25 -0.57529
26 -0.99498
27 3.95229
28 -0.37182
29 -0.84187
30 0.83796
31 0.25449
32 -0.06097
Distance: 100.000000
# Gi*(d)
1 -0.35877
2 -0.56799
3 1.27146
4 0.03060
5 0.65710
6 0.28850
7 0.22455
8 0.91189
9 0.56230
10 0.84063
11 0.20650
12 0.56230
13 -0.22102
14 0.06935
15 -1.48652
16 0.27522
17 -1.46440
18 -1.45026
19 -0.19244
20 0.05359
21 -1.59361
22 0.20424
23 -0.35877
24 -0.81246
25 -1.12633
26 -1.35693
27 3.95229
28 -0.37182
29 -1.76916
30 0.38227
31 -0.40318
32 -0.15433
Distance: 150.000000
# Gi*(d)
1 -0.72769
2 -0.61369
3 1.21299
4 -0.73709
5 0.28850
6 0.44533
7 1.89823
8 1.17137
9 1.74185
10 -0.40733
11 0.03118
12 0.19677
13 0.86130
14 2.57943
15 -1.27227
16 2.14461
17 -1.09643
18 -1.64603
19 2.34872
20 -0.23837
21 -1.71618
22 0.69550
23 -1.37782
24 -1.26129
25 -1.07759
26 -1.27227
27 1.80685
28 2.04694
29 -1.55458
30 -0.47802
31 0.19809
32 -1.06122
Distance: 200.000000
# Gi*(d)
1 -1.60199
2 0.97606
3 0.99883
4 -1.74106
5 0.33635
6 0.56721
7 1.68767
8 0.71302
9 1.48808
10 0.25014
11 -0.74365
12 1.74185
13 1.02729
14 2.07760
15 -1.49910
16 1.38907
17 -1.52031
18 -0.85436
19 2.74986
20 0.19228
21 -1.83500
22 0.33635
23 -1.09888
24 -2.15949
25 -0.87391
26 -1.82133
27 1.40557
28 0.86386
29 -1.71076
30 -0.57175
31 -0.45067
32 -1.75313
Distance: 250.000000
# Gi*(d)
1 -0.72829
2 0.65445
3 0.95940
4 -1.49865
5 0.50154
6 -0.24102
7 2.13038
8 0.76704
9 1.40557
10 -0.33502
11 -0.71390
12 1.48808
13 0.99883
14 2.43843
15 -1.89907
16 1.40557
17 -2.17783
18 -0.87391
19 2.82523
20 -0.12649
21 -1.99650
22 0.74490
23 -0.59005
24 -2.20728
25 -1.07311
26 -1.66895
27 1.49201
28 0.89900
29 -1.15890
30 0.76238
31 -0.57532
32 -0.67862
Distance: 300.000000
# Gi*(d)
1 -0.41366
2 0.96351
3 1.56281
4 -0.85436
5 0.57030
6 -0.26637
7 2.33549
8 1.93695
9 1.48872
10 -0.65520
11 -0.45403
12 1.40557
13 0.99883
14 1.68321
15 -1.60165
16 1.40557
17 -0.36352
18 -0.94853
19 2.10932
20 -0.19851
21 -2.59936
22 0.57030
23 -0.25629
24 -2.03640
25 -0.94853
26 -0.01566
27 1.54522
28 0.21334
29 -1.29619
30 -1.08038
31 -1.04677
32 -1.04106
References
Ord, J.K. and Getis, A., (1995) Local Spatial Autocorrelation Statistics: Distribution Issues and an Application, Geographical Analysis, 27(4): 286-306
National Cancer Institute Biometry Research Group Datasets
http://dcp.nci.nih.gov/bb/datasets.html