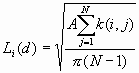 [1]
[1]Local K-Function
The local K-function was developed by Getis (1984). It is similar to the global K-function in analysis, but differs in that the local K-function only considers those pairs of points having a given point i as one of its members.
Input
Analysis
Like the K-function, the Local K-function is a test of the hypothesis of CSR, and the expected value of Li(d) is d. Again, the confidence envelope is generated by performing a specified number of simulations. If for any distance the observed Li(d) falls outside the confidence envelope, the hypothesis of CSR can be rejected at the appropriate significance level. An observed Li(d) below the envelope indicates that the points are dispersed about point i for distance d. Conversely, an observed Li(d) above the envelope indicates that the points are clustered about point i for distance d.
Formula
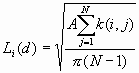 [1]
[1]
where:
A is the study area,
N is the number of points
d is the distance
![]() is the number of points within distance d of point i
is the number of points within distance d of point i
![]() is the weight, which includes boundary corrections. The weights are the same as those used in the K-function.
is the weight, which includes boundary corrections. The weights are the same as those used in the K-function.
Output
The output file includes the input data file, the total number of points, the minimum and maximum X,Y coordinates, the size of the study area, maximum search distance, number of intervals, and the permutations used in creating the confidence envelope. For each specified distance the following table is printed.
|
Points |
Observed Li(d) |
Li(d)-d |
|
1 2 : |
Example
For this example we will consider the same data that is used for the Knox statistic example. We are examining cases of an infectious disease during an outbreak. The data includes the X and Y coordinates in meters of each case. A sample of the input data file is shown in Table 1.
Table 1: Input File
X Y Z
138902 58938 1 137625 59262 1 138431 58633 1 138637 58586 1 137738 58994 1 . . . . . . . . . 139641 61019 1
The disease is transmitted by a vector that is believed to operate over short distances (less than 35 meters). We will use 35m as our maximum distance of study for this test. A sample of the output is shown in Table 2.
Table 2: Sample Output File
The input data file: cases.dat
The total number of points: 294
The minimum x coordinate: 135794.000000
The maximum x coordinate: 141456.000000
The minimum y coordinate: 55984.000000
The maximum y coordinate: 61643.000000
The total area: 32041258.000000
The maximum search distance: 35.000000
The step size: 35.000000
The number of permutation for significance envelope:99
Distance: 35.00 Minimum Li(d): 0.000 Maximum Li(d): 263.852
Point# Observed Li(d) Li(d)-d
1 0.000 -35.000
2 186.572 151.572
3 417.187 382.187
That output file shows that there were a total of 294 cases, and the complete output file contains a Li(d) value for each point. We have chosen to discuss the output of three key points. These points are shown in the output as points 1,2, and 3.
The Li(d) for point #1 is 0.00. This indicates that there are no other points within 35m of point #1. We can see that this Li(d) value is equal to the minimum Li(d) on the confidence envelope. This indicates that we expect a number of cases not to have a neighboring case within 35m.
The Li(d) for point #2 is 186.57. This value is well within the confidence envelope. Although there are one or more cases within 35m of point #2, there is not significant clustering of cases around point #2.
The Li(d) for point #3 is 417.187. This value is well above the maximum Li(d), and we reject the null hypothesis of a CSR distribution. Recall that a value above the confidence envelope indicates clustering. We can conclude that there is a significant clustering of cases around point #3.
We can see from these three examples that the local K-function gives us an idea of the distribution of points around each point individually.
References
Getis, A, (1984), Interaction Modeling Using Second-order Analysis. Environment and Planning A 16: 173-183
Morrison, Amy C., Getis, Arthur, Santiago, Marilyn, Rigau-Perez, Jose G., and Reiter, Paul (1998). Exploratory Space-Time Analysis of Reported Dengue Cases During an Outbreak in Florida, Puerto Rico, 1991-1992. Am J Trop Med Hyg 58(3): 287-298