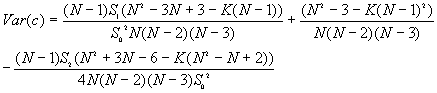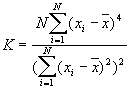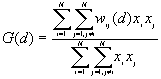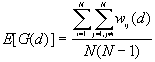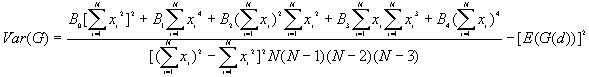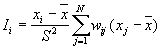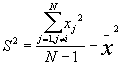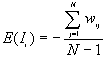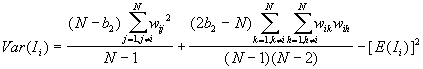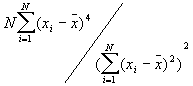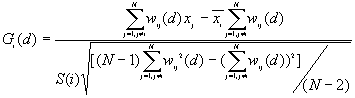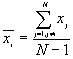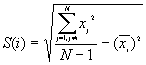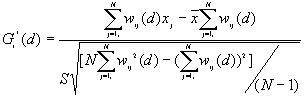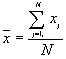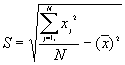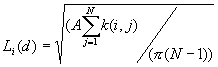POINT PATTERN ANALYSIS (PPA)
by
DongMei Chen and Arthur Getis
Department of Geography
San Diego State University
San Diego, CA 92118-4493
dchen@rohan.sdsu.edu
arthur.getis@sdsu.edu
May 12, 1998
TABLE OF CONTENTS
Introduction
Routines
- Basic descriptive statistics
- Nearest neighbor analysis
- Refined nearest neighbor analysis
- K-function (or second order analysis)
- Weighted K-function
- Cluster: Knox space-time
- Join-count statistics
- Global Moran's I
- Global Geary's c
- General Getis-Ord's G(d)
- Local Moran's
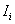
- Local
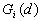
- Local
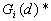
- Local K-Function
POINT PATTERN ANALYSIS (PPA)
Introduction
This Point Pattern Analysis (PPA) software package is written and compiled in C and is used to describe and help analyze point patterns. It consists of 14 different analysis routines. These represent a variety of basic descriptive statistics and include: nearest neighbor analysis, refined nearest neighbor analysis, K-function, weighted K-function, space-time Knox, Join-Count statistics, Global Moran's I and Geary's c, general Getis-Ord's G, local Moran's 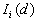 , local
, local 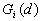 and
and 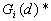 , and local K-function. This manual contains a brief description of each analysis as well as input and output information.
, and local K-function. This manual contains a brief description of each analysis as well as input and output information.
This PPA package can be run in both DOS (or a DOS program in WINDOWS) and UNIX. The memory requirement for running PPA depends on the size of the data set.
In order to run this package, you need to copy the executable files ppa.exe (for DOS), or ppa (for UNIX) to a new directory that you want to work on and then type
ppa
followed by a Return. This will clear the screen and a welcome message will appear. Press Enter key, then choose the desired routine.
All routines (except Join-Count) are designed for data in three columns X, Y and Z, where X, Y are coordinates, and Z represents the value at site X, Y or time. If all Z values are weighted similarly, that is, they are to be evaluated as single, unweighted points, a column of 1s should make up the Z column. If a weight matrix file is used, it should be organized as an N by N matrix in the order of your input data.
All the results will appear on the screen (UNIX) and be saved to a file that you name. PPA will empty this file before it saves any new output.
Instruction
- Call for directory ppa
- Type ppa
- Press Enter key to begin
- Choose routine member
- Enter input data
- Enter output data file name (any name will do)
- To continue, enter 1 (computing will now take place)
- Enter 0 to quit
- Call for output file
1. Basic Descriptive Statistics (BDS)
In this option, the minimum, maximum, mean, standard deviation, skewness, and kurtosis are calculated. Standard deviation measures dispersion from the mean, skewness measures the extent to which the bulk of the values in a distribution are concentrated to one side or the other of the mean, kurtosis measures the extent to which values are concentrated in one part of a frequency distribution. The formulas for these parameters are the following:
Mean 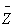 =
=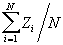
Standard Deviation Std(Z) = 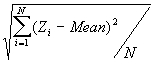
Skewness = 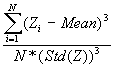
Kurtosis = 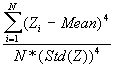
where N is the total number of points.
Input
The data file contains N rows of X, Y coordinates and Z values.
Output
The output lists the total number of points in the files, the minimum, maximum, mean, standard deviation, skewness and kurtosis values for X, Y and Z.
2. Nearest Neighbor Analysis
Nearest neighbor analysis examines the distances between each point and the closest point to it, and then compares these to expected values for a random sample of points from a CSR (complete spatial randomness) pattern.
Formula:
a) The mean nearest neighbor distance
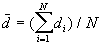 [1]
[1]
where N is the number of points. 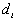 is the nearest neighbor distance for point i.
is the nearest neighbor distance for point i.
b) The expected value of the nearest neighbor distance in a random pattern
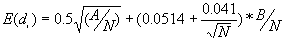 [2]
[2]
where A is the area and B is the length of the perimeter of the study area.
c) The variance
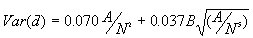 [3]
[3]
Equations [2] and [3] contain a correction factor to account for the boundary effect based on Donnelly (1978).
Input
You'll be asked to enter the input data file, which should contain N rows of X, Y coordinates, and Z values. Make Z values all equal to 1 representing points.
Output
The output file lists a) the input data file, b) the total number of points, c) the minimum and maximum of the X, and Y coordinates, d) the size of study area, e) the observed mean nearest neighbor distance, f) the expected average nearest neighbor distance, g) the variance, and h) Z statistic (standard normal variate). A negative Z score indicates clustering; a positive score means dispersion or evenness.
Limitation
Equations [2] and [3] cannot be used for irregularly shaped study areas. In this program, the study area is a regular rectangle or a square. A is calculated by (Xmax - Xmin) * (Ymax - Ymin).
3. Refined Nearest Neighbor Analysis
Refined nearest neighbor analysis involves comparing the complete distribution function of the observed nearest neighbor distances,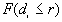 , with the distribution function of the expected nearest neighbor distances for CSR,
, with the distribution function of the expected nearest neighbor distances for CSR, 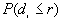 . The program finds the largest absolute difference
. The program finds the largest absolute difference 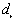 , and tests for significance based on a Monte Carlo test.
, and tests for significance based on a Monte Carlo test.
Formula
a) 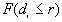 is obtained by taking the nearest neighbor distances,
is obtained by taking the nearest neighbor distances, 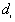 , and the nearest distances to study boundary,
, and the nearest distances to study boundary, 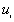 , for each point i. The program ranks
, for each point i. The program ranks 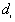 from the smallest to the largest. For every distance of interest,
from the smallest to the largest. For every distance of interest, 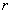 , the program counts the number of points
, the program counts the number of points 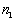 for which
for which 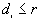 , and the number of points
, and the number of points 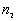 for which
for which 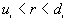 . The observed proportion of the nearest neighbor distances less than or equal to some chosen distance
. The observed proportion of the nearest neighbor distances less than or equal to some chosen distance 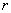 is decided by equation [1].
is decided by equation [1].
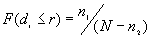 [1]
[1]
Where N is the total number of points.
b) The proportion of expected nearest neighbor distances less than or equal to r for an unbounded CSR pattern is:
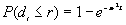 [2]
[2]
Where:
e is the mathematical constant 2.718283....
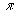 is the mathematical constant 3.141593....
is the mathematical constant 3.141593....
r is the specified distance
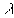 is the estimated point density (N/A)
is the estimated point density (N/A)
c)
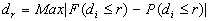
Where Max | | means the largest absolute value obtained for corresponding values of r.
Input
You'll be asked to enter the input data file, which should contain rows of X, Y coordinates representing points, and Z values made up of 1s.
Output
The output file includes three parts: the first part lists a) the input data file, b) the total number of points, c) the minimum and maximum of X, and Y coordinates, and d) the size of study area.
The second part is a table of the following form:
|
r
(distance) |
Observed number of points (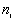 ) for which ) for which 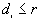 |
Observed number of points (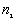 ) for which ) for which 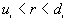 |
Observed proportion 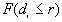 |
Expected proportion 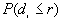 |
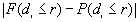
|
|
:
:
: |
|
|
|
|
|
If for each r, 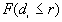 >
>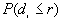 , a clustered pattern is indicated, whereas
, a clustered pattern is indicated, whereas 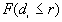 <
<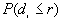 indicates a regular pattern of points.
indicates a regular pattern of points.
The last part shows 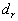 (the largest absolute value obtained for
(the largest absolute value obtained for 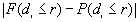 ), r (the distance for
), r (the distance for 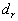 ), and its significance. If F is greater than P, then clustering is implied.
), and its significance. If F is greater than P, then clustering is implied.
Limitation
In this program, the study area is a regular rectangle or a square. The area is calculated by (Xmax - Xmin) * (Ymax - Ymin).
4. K-Function
K-Function is also called second-order analysis to indicate that the focus is on the variance, or second moment, of interevent distances. It considers all combinations of pairs of points. It compares the number of observed pairs with the expectation at all distances, taking into consideration the density of points, the borders, and the size of the sample.
Formula
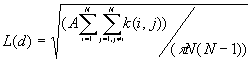 [1]
[1]
Where:
A is the size of study area,
N is the number of points,
d is the distance,
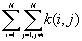 is the number of j points within distance d of all i points. k(i, j) is the weight, which is estimated by
is the number of j points within distance d of all i points. k(i, j) is the weight, which is estimated by
a) If no edge corrections,
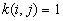 in case
in case 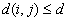
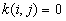 otherwise
otherwise
b) If a point i is closer to one boundary than it is to a point j, the border correction is employed.
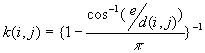
where e is the distance to the nearest edge.
c) If a point i is closer to two right angle boundaries than it is to a point j, the weighting formula is
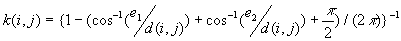
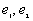 are the distances to the nearest vertical and horizontal borders respectively.
are the distances to the nearest vertical and horizontal borders respectively.
The expectation for a CSR pattern of L(d) is d.
Input
1. The input data file, which should include the X, Y coordinates of points, and Z values ( a column of 1s).
2. The maximum distance (dmax) that you want. Usually a statistically unbiased maximum distance is less than the circumradius of the study area, or one-half the lesser of the length or the width of a rectangular study area.
3. The number of increments.
4. The number of permutations for creating the confidence envelope.
5. Output file.
Output
This program calculates the distances d(i,j) between all combinations of two points, and calculate the k(i, j) for all pairs, and then calculate the L(d) for all d. The program will randomly generate the N points in the whole study area M times, and get the minimum and maximum of L(d) for the envelope. The output lists the input data file, the total number of points, the minimum and maximum of x and y coordinates, the size of study area, and the following table
|
distance d |
Observed L(d) |
L(d) - d |
Minimum L(d) |
Maximum L(d) |
|
:
:
: |
|
|
|
|
Limitation
The boundary correction formulas used here are inappropriate for irregular borders. In this program we assume the study area is a rectangle or a square.
5. Weighted K-Function
The weighted K-function was developed by Getis (1982) based on the K-Function. It considers both location and the value of a point. The statistical test is based on the assumption of CSR, and performed on independent simulations of all values in fixed locations of the study area.
Formulas
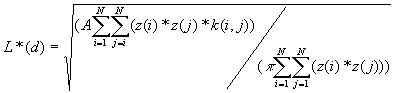 [1]
[1]
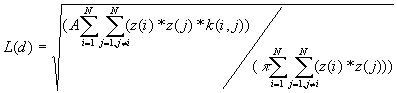 [2]
[2]
Equation [1] includes the i points' interaction with all points, including itself while in [2] i does not equal j.
Where A is the size of the study area,
z(i) is the weight of the point i,
k(i, j) is the border correction value, the same as that defined for the K-Function.
Input
1. The input data file, contains rows of the (x, y) coordinates and the z values of points.
2. The maximum distance (dmax); usually statistical unbiased maximum distance is less than the circumradius of the study area, or one-half the lesser of the length or the width of a rectangular study area.
3. The number of increments.
4. The number of permutations for creating the confidence envelope.
5. Output file.
Output
This program calculates the distances d(i,j) between all combinations of two points, and calculates the k(i, j) for all pairs, and then calculates the L(d) and L*(d) for all d. For the confidence envelope, the program will randomly assign the Z values of each point to the N points M times, and find the minimum and maximum of L(d) and L*(d). The output lists the input data file, the total number of points, the minimum and maximum of X, and Y coordinates, the size of study area, and the following two tables showing L(d) and L*(d) respectively:
|
distance (d) |
Observed L(d) |
Minimum L(d) |
Maximum L(d) |
|
:
:
: |
|
|
|
|
distance (d) |
Observed L*(d) |
Minimum L*(d) |
Maximum L*(d) |
|
:
:
: |
|
|
|
Limitation
The boundary correction formulas used here are inappropriate for irregular borders. In this program we assume the study area is a regular rectangle or a square.
6. Knox Statistic for Space-Time Clustering
The Knox approach is used to test whether there is a significant cluster during a defined distance and time period. First it counts the number of point pairs as either close or distant in space and /or time, then calculates the P-value.
Formula
For a certain distance d and time period t, the Knox statistic calculates the following number:
d(i, j) is the distance of point i and j,
t(i, j) is the time interval of point i and j,
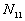 : the number of point pairs (i, j) with d(i, j)
: the number of point pairs (i, j) with d(i, j) 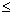 d, and t(i, j)
d, and t(i, j) 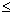 t,
t,
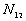 : the number of point pairs (i, j) with d(i, j)
: the number of point pairs (i, j) with d(i, j) 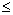 d, and t(i, j) > t,
d, and t(i, j) > t,
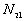 : the number of point pairs (i, j) with d(i, j) > d, and t(i, j)
: the number of point pairs (i, j) with d(i, j) > d, and t(i, j) 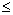 t,
t,
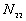 : the number of point pairs (i, j) with d(i, j) > d, and t(i, j) > t,
: the number of point pairs (i, j) with d(i, j) > d, and t(i, j) > t,
N is the total number of point pairs (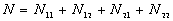 )
)
The P-value is:
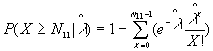
where 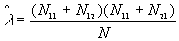 ,
,
Input
1. Input data file, which should record X, Y coordinates of points and T the times attached to each points (time elapsed in days, or years or minutes, etc).
2. The time interval.
3. The distance interval.
4. Output file.
Output
1. The input data file name,
2. The total number of points,
3. The minimum and maximum of X, and Y coordinates, and time.
4. The time and distance intervals.
5. The number of point pairs tabulated as
| |
t(i, j) <= t |
t(i, j) > t |
Space only |
|
d(i, j) <= d |
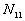
|
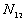
|
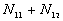
|
|
d(i, j) > d |
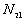
|
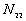
|
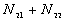
|
|
Time only |
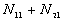
|
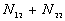
|
N |
6. EN11 is the expected value of 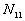 .
.
6. The P-value. Low P-values (e.g., 0.01) represent significant time-space clustering.
7. Join-Count Statistics for Spatial Autocorrelation (Free sampling model)
Join-Count statistics are the simplest measure of spatial autocorrelation. They are used for a binary variable ( 1 or 0 ). The two values of the variable are referred to as "black" (B) and "white" (W). A join links two neighboring areas. So the possible types of joins are black-black (BB), black-white (BW), and white-white (WW). Join counts are counts of the numbers of BB, BW, and WW joins in the study area, and these numbers are compared to the expected numbers of BB, BW and WW joins under the null hypothesis of no spatial autocorrelation.
Formulas
The observed number of BB, BW and WW joins are given by
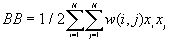 [1]
[1]
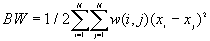 [2]
[2]
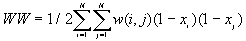 [3]
[3]
Where 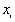 is the binary value, 1 for black, 0 for white,
is the binary value, 1 for black, 0 for white,
w(i, j) is the binary weight, 1 if two areas are contiguous, 0 otherwise.
For different assumptions about the data, the theoretical expressions for E(BB), E(BW) and E(WW) will vary. Under the free sampling model, the expected BB, BW and WW are:
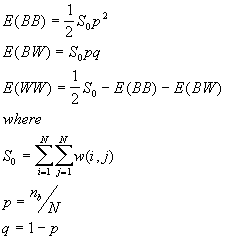
and 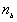 is the number of areas with B values.
is the number of areas with B values.
The variances are
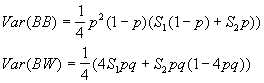
Where
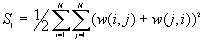
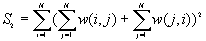
Input
1. Input data file, which records the binary value for each area.
2. Input weight matrix file, which is an N by N weight matrix with 1 for contiguous areas, 0 otherwise.
3. Output file.
Output
1. The total number of areas,
2. The total number of black areas,
3. The total number of white areas,
4. The observed number of BB, BW and WW joins,
5. The expected number of BB, BW, and WW joins,
6. The variance of BB, BW joins,
7. The z-statistics of BB, BW joins.
8. Global Moran's I
9. Global Geary's c
Moran's I and Geary's c are well known for testing for spatial autocorrelation. They represent two special cases of the general cross-product statistic that measures spatial autocorrelation. Moran's I is produced by standardizing the spatial autocovariance by the variance of the data using a measure of the connectivity of the data. Geary's c uses the sum of squared differences between pairs of data values as its measure of covariation.
Formula
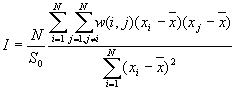 [1]
[1]
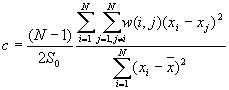 [2]
[2]
Where 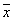 is the mean of
is the mean of 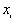 ,
, 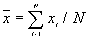 ,
,
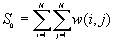
The expected value of Moran's I is -1/(N-1). Values of I that exceed -1/(N-1) indicate positive spatial autocorrelation, in which similar values, either high values or low values, are spatially clustered. Values of I below -1/(N-1) indicate negative spatial autocorrelation, in which neighboring values are dissimilar.
The theoretical expected value for Geary's c is 1. A value of Geary's c of less than 1 indicates positive spatial autocorrelation, while a value larger than 1 points to negative spatial autocorrelation.
The variances of I and c will vary for different assumptions about the data. Under the randomization assumption, the variance of I and c are
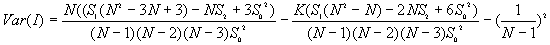
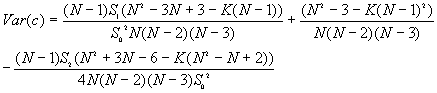
where
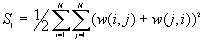
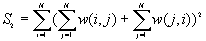
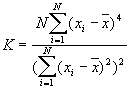
The values of Moran's I and Geary's c depend on the w(i,j), which are specified by the spatial weighting scheme chosen. In this program, two weighting schemes can be selected:
a. The w(i, j) are equal to the values in the input N by N matrix taken from the spatial weight matrix file that the user has prepared.
b. The 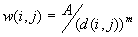 , where A is a constant (usually set at 1), d(i,j) is the distance between the ith and jth points; m is a parameter representing the friction of distance selected a priori.
, where A is a constant (usually set at 1), d(i,j) is the distance between the ith and jth points; m is a parameter representing the friction of distance selected a priori.
In order to evaluate spatial trends in the pattern, sometimes it is necessary to identify spatial autocorrelation at several levels of spatial separation ( in the form of a spatial correlogram), such as for different orders or distances of neighboring points. In this program, two different correlograms for I and c are available, one is the change by bands (Figure 1a), and the other is by distance increments (Figure 1b).
. .
(a): Bands (b): Increments
Figure 1: in (a), points found in the band represented by the shadowed concentric circle are related to the ith point shown at the center. The correlogram shows the relationship of points in this band and further bands to each of the i points taken together. In (b), points found in the shadowed region are related to the ith point at the center. The correlogram shows the cumulative relationship of points at a series of distances from the i points.
Input for I and/or c
1. Input data file contains the X, Y coordinates and the value at each point.
2. Input whether you have a spatial weight matrix file.
3. Select the weighting scheme. If you select b, you'll be asked to enter the A and m parameter.
4. Select whether you want to calculate a single I (or c), or correlogram. If correlogram is selected, you will be asked to enter the maximum distance (dmax), steps you want (nstep), and whether by bands or increments.
Output for Moran's I
The output depends on the your input. For each distance range, this program will output
a. the total number of points,
b. observed I,
c. expected I,
d. the variance,
e. z value
Output for Geary's c
The output depends on the your input. For each distance range, this program will output
a. the total number of points,
b. observed c,
c. the variance,
d. z value
10. General G(d) statistic
The G statistics were developed by Getis and Ord (1992). It is a multiplicative measure of overall spatial association of values which fall within a critical distance of each other. It can only be computed for positive variables.
Formula
For a chosen critical distance d, G(d) is
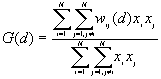
where 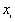 is the value of ith point,
is the value of ith point,
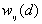 is the weight for point i and j for distance d.
is the weight for point i and j for distance d.
The expected mean value of G(d)
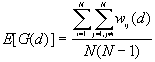
The variance of G(d)
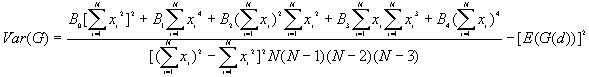
where
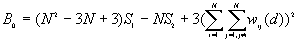
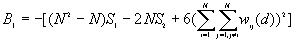
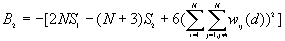
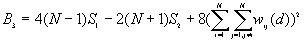
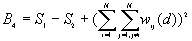
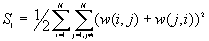
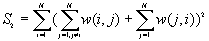
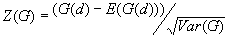
A positive z-value for G(d) indicates spatial clustering of high values, and a negative z-value indicates spatial clustering of low values.
Input
1. Input data file, which records the X, Y coordinate and the value of points.
2. The maximum distance.
3. The number of increments.
4. The output file.
Output
1. The number of points.
2. The distance and its corresponding G(d), expected G(d), Variance and Z value (standard variates).
11. Local Spatial Autocorrelation Statistics
Local spatial autocorrelation statistics are observation-specific measures of spatial association. They focus on the location of individual points, and allow for the decomposition of global or general statistics into the contribution by each individual observation. It can be used to detect the local spatial clustering around an individual location, spatial nonstationarity, the outline of spatial regimes, especially in cases where global statistics may fail to detect these patterns, or where a single measure of global association may contribute little meaning
Local Moran's 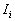
According to Anselin (1995), a local Moran statistic for a point i is defined as
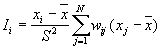
where
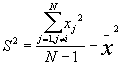
For a randomization hypothesis, the expected value is
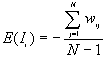
The variance is as
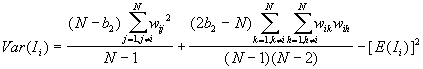
where
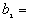
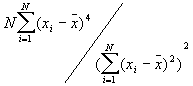
Note: this statistics is calculated for bands only in this package.
Input
1. Input data file, which record the X, Y coordinates and the values of points.
2. The distance used.
3. The parameter m used in weighting scheme (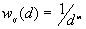 ).
).
4. Output file.
Output
1. The number of points,
2. The distance used,
3. The local Moran's Ii, the expected value, variance, z-value for each point.
12. Local 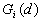
13. Local 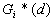
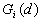 and
and 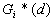 were developed by Ord and Getis (1995). They indicate the extent to which a location is surrounded by a cluster of high or low values. The
were developed by Ord and Getis (1995). They indicate the extent to which a location is surrounded by a cluster of high or low values. The 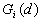 statistic excludes the value at i from the summation while the
statistic excludes the value at i from the summation while the 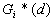 includes the value at i. Positive
includes the value at i. Positive 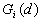 or
or 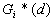 indicate spatial clustering of high values, whereas negative
indicate spatial clustering of high values, whereas negative 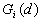 or
or 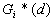 indicate spatial clustering of low values.
indicate spatial clustering of low values.
Formulas
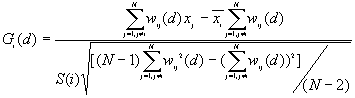
where
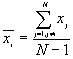
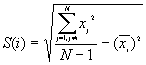
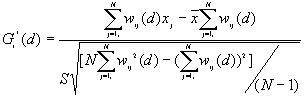
Where
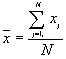
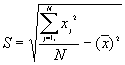
Both 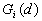 and
and 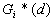 are asymptotically normally distributed as d increases.
are asymptotically normally distributed as d increases.
Under the null hypothesis that there is no association, the expectation value is 0, the variance is 1. If the underlying data are normally distributed, we can consider their values as standard variates.
Input
1. Input data file,
2. The maximum distance,
3. The number of increments,
4. Output file.
Output
1 The number of points,
2 The distance used,
3 The 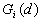 or
or 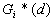 for each point.
for each point.
|
point |
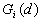 or or 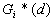
|
|
: |
|
14.
Local K-Function
The local K-Function was developed by Getis (1984). It is similar to the global K-function analysis, but differs in that the local K-function only considers those pairs of points having as one of its members a given point i.
Formula
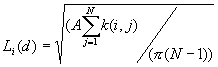
Where:
A is the size of study area,
N is the number of points,
d is the distance,
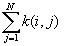 is the summation over all points that are within distance d of point i, and it includes a boundary correction that is the same as that in the K-function program.
is the summation over all points that are within distance d of point i, and it includes a boundary correction that is the same as that in the K-function program.
Input
1. The input data file includes the X, Y coordinates of points, and Z values ( assign 1s).
2. The maximum distance (dmax) that you want, usually statistically unbiased maximum distance is less than the circumradius of the study area, or one-half the lesser of the length or the width of a rectangle study area.
3. The number of increments.
4. The number of permutations for creating the confidence envelope.
5. Output file.
Output
This program calculates the number of pairs of points between i and all points within d, and calculate the k(i, j), and then calculate the Li(d) for each i and d. The program will randomly generate the N points in the whole study area M times, and found the minimum and maximum of L(d) for the envelope. The output lists the input data file, the total number of points, the minimum and maximum of X and Y coordinates, and the size of the study area. For each distance, the output lists the distance, the minimum and maximum L(d) , and the following table.
|
Points |
Observed Li(d) |
Li(d) - d |
|
1
2
: |
|
|
References
Anselin, L. (1995) SpaceStat Tutorial: A Workbook for Using SpaceStat in the Analysis of Spatial Data. NCGIS, Santa Barbara.
Anselin, L. (1995) The Local Indicators of Spatial Association – LISA, Geographical Analysis, 27: 93-115.
Boots, B.N. and Getis, A (1988) Point Pattern Analysis, Sage: Newbury Park, CA.
Cliff, A.D. and Ord, J.K (1973) Spatial Autocorrelation, Pion: London.
Cliff, A.D. and Ord, J.K (1981) Spatial Processes: Models and Applications, Pion:London.
Cressie, N.A. (1991) Statistics for Spatial Data, John Wiley: Chichester.
Diggle, P. and Chetwynd, A.G (1991) Second-order analysis of spatial clustering, Biometrics 47:1155-1163.
Gatrell, A.C, Bailey, T.C., Diggle, P and Rowlingson, B.S.(1996) Spatial Point Pattern Analysis and its Application in Geographical Epidemiology, Trans. Inst Br Geogr NS 2: 256-274.
Getis, A (1984) Interaction Modeling Using Second-order Analysis. Environment and Planning A 16: 173-183.
Getis, A and J. Franklin (1987) Second-order Neighborhood Analysis of Mapped Point Patterns. Ecology, 68(3): 473-477.
Getis, A and Ord, J.K (1992) The Analysis of Spatial Association by Use of Distance Statistics, Geographical Analysis, 24: 189-206.
Getis, A and Ord, J.K (1996) Local Spatial Statistics: An Overview. In Spatial Analysis: Modeling in a GIS Environment, edited by P. Longley and M. Batty. John Wiley & Sons: New York.
Ord, J.K and Getis, A., (1995) Local Spatial Autocorrelation Statistics: Distribution Issues and an Application, Geographical Analysis, 27(4): 286-306.
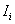
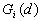
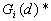
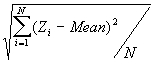
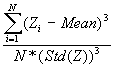
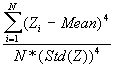
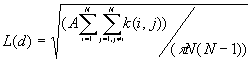 [1]
[1]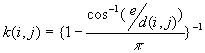
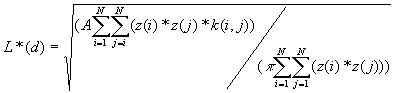 [1]
[1]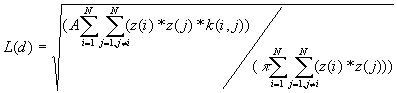 [2]
[2]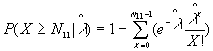
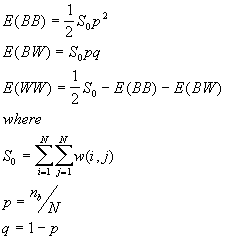
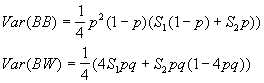
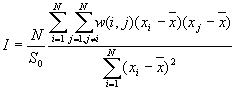 [1]
[1]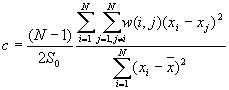 [2]
[2]