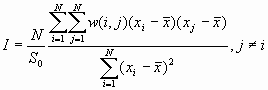 [1]
[1]Global Moranís I and Global Gearyís c
Moranís I and Gearyís c are well known tests for spatial autocorrelation. They represent two special cases of the general cross-product statistic that measures spatial autocorrelation. Moranís I is produced by standardizing the spatial autocovariance by the variance of the data. Gearyís c uses the sum of the squared differences between pairs of data values as its measure of covariation. Both of these statistics depend on a spatial structural specification such as a spatial weights matrix or a distance related decline function.
Input
Analysis
The expected value of Moranís
I is -1/(N-1). Values of I that exceed -1/(N-1) indicate positive spatial autocorrelation, in which similar values, either high values or low values are spatially clustered. Values of I below -1/(N-1) indicate negative spatial autocorrelation, in which neighboring values are dissimilar.The theoretical expected value for Gearyís c is 1. A value of Gearyís c less than 1 indicates positive spatial autocorrelation, while a value larger than 1 points to negative spatial autocorrelation.
Formula
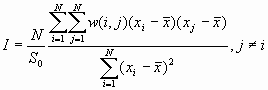 [1]
[1]
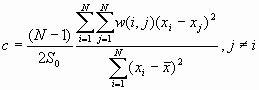 [2]
[2]
where ![]() is the mean of
is the mean of ![]() ,
, ![]() ,
, ![]() , and w(i,j) is the connectivity spatial weight between I and j.
, and w(i,j) is the connectivity spatial weight between I and j.
The variances of I and c will differ according to the data model employed. PPA uses a randomization assumption. Under a randomization assumption, the variances of I and c are shown below.
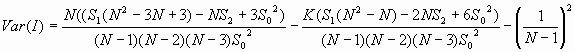
![]()
![]()
where
![]()
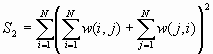
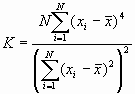
The values of Moranís
I and Gearyís c depend on the w(i , j), which are specified by the spatial weighting scheme chosen. In this program, two weighting schemes can be selected:, where
d(i , j) is the distance between the ith and the jth points; m is a parameter representing the friction of distance selected a priori; A is usually set equal to 1.In order to evaluate spatial trends in the pattern, sometimes it is necessary to identify spatial autocorrelation at several levels of spatial separation (in the form of a spatial correlogram). In this program, two different correlograms for I and c are available. One type is autocorrelation by bands (Figure 1a) and the other is by cumulative distance increments (Figure 1b).
Figure 1: Correlograms
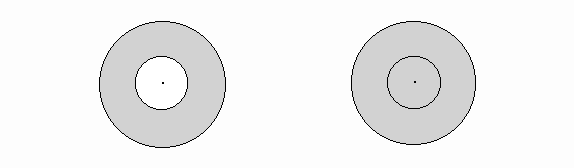
a) bands b) increments
In a, points found in the band represented by the shaded concentric circle are related to the ith point shown in the center. The correlogram shows the relationship of points in each band (from near to far). In b, points found in the shadowed region are related to the ith point at the center. In this case, the correlogram shows the cumulative relationship of points at a series of distances from the i points.
Output for Moranís I
For each distance range, the program will output
Output for Gearyís c
For each distance range, the program will output
Example
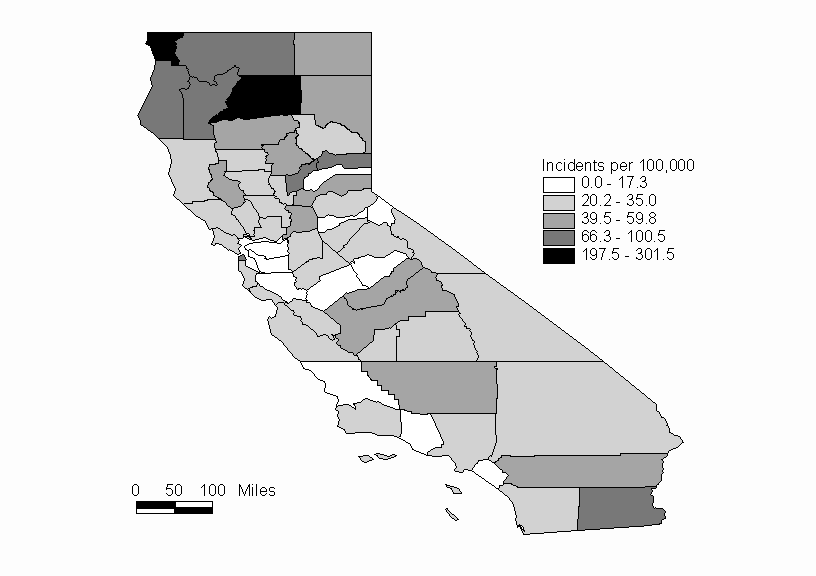
For this example we will consider the distribution of hepatitis rates for the counties of California. The data are taken from the Department of Health Services of the State of California (1999). The rates are given as cases per 100,000 population, and are calculated by using 1998 data over the average population from 1995-1997. The data are shown in Table 1. A map showing the hepatitis rates by county is shown in Figure 2.
Table 1: Reported Hepatitis Rates of California Counties
|
County |
X |
Y |
Rate |
|
Alameda |
195 |
500 |
14.4 |
|
Alpine |
318 |
560 |
0 |
|
Amador |
265 |
550 |
12.1 |
|
Butte |
220 |
630 |
52.9 |
|
Calaveras |
280 |
530 |
22.6 |
|
Colusa |
195 |
598 |
23.8 |
|
Contra Costa |
192 |
515 |
12.5 |
|
Del Norte |
100 |
790 |
301.5 |
|
El Dorado |
260 |
580 |
32 |
|
Fresno |
320 |
425 |
53.9 |
|
Glenn |
180 |
630 |
35 |
|
Humboldt |
90 |
705 |
100.5 |
|
Imperial |
648 |
56 |
66.3 |
|
Inyo |
450 |
403 |
29.3 |
|
Kern |
396 |
256 |
41.2 |
|
Kings |
315 |
380 |
21.9 |
|
Lake |
155 |
597 |
39.5 |
|
Lassen |
270 |
710 |
59.2 |
|
Los Angeles |
436 |
168 |
21 |
|
Madera |
315 |
455 |
45 |
|
Marin |
175 |
510 |
20.2 |
|
Mariposa |
305 |
485 |
10.4 |
|
Mendocino |
125 |
602 |
27.5 |
|
Merced |
285 |
470 |
16.6 |
|
Modoc |
265 |
765 |
59.8 |
|
Mono |
380 |
515 |
31.6 |
|
Monterey |
212 |
415 |
26.6 |
|
Napa |
185 |
545 |
23.8 |
|
Nevada |
255 |
610 |
13.8 |
|
Orange |
468 |
112 |
17.3 |
|
Placer |
270 |
595 |
50.8 |
|
Plumas |
272 |
660 |
34.6 |
|
Riverside |
600 |
120 |
46.5 |
|
Sacramento |
235 |
548 |
43.1 |
|
San Benito |
220 |
430 |
25 |
|
San Bernadino |
584 |
216 |
33.7 |
|
San Diego |
544 |
52 |
22.4 |
|
San Fransisco |
185 |
503 |
78.2 |
|
San Joaquin |
236 |
520 |
30.5 |
|
San Luis Obispo |
272 |
260 |
11.8 |
|
San Mateo |
190 |
490 |
26.6 |
|
Santa Barbara |
300 |
200 |
24.4 |
|
Santa Clara |
202 |
475 |
14.8 |
|
Santa Cruz |
200 |
450 |
27.8 |
|
Shasta |
197 |
712 |
197.5 |
|
Sierra |
275 |
630 |
78.4 |
|
Siskiyou |
180 |
782 |
75.9 |
|
Solano |
192 |
540 |
23.6 |
|
Sonoma |
170 |
535 |
24.6 |
|
Stanislaus |
265 |
491 |
26.8 |
|
Sutter |
210 |
590 |
32.6 |
|
Tehama |
193 |
680 |
58.3 |
|
Trinity |
140 |
702 |
75 |
|
Tulare |
365 |
385 |
30.3 |
|
Tuolumne |
303 |
515 |
20.7 |
|
Ventura |
372 |
176 |
16.2 |
|
Yolo |
205 |
570 |
30.6 |
|
Yuba |
228 |
604 |
79.8 |
Figure 2: Hepatitis Rates of California Counties in 1998 (per 100,000 pop.)
For this example we will use the following weighting scheme:
![]() ,thus A = 1 and m = 2
,thus A = 1 and m = 2
Both Moranís I and Gearyís c results are shown in Table 2. The Moranís I and Gearyís c statistics are calculated for 50-mile increments from 50 to 250 miles. For each of these increments, the Gearyís c is less than 1, and the Moranís I is greater than the expected value. These results indicate that there is positive spatial autocorrelation. However, none of the Z-values are significant at the a =0.05 level, and we can not reject the null hypothesis of a random distribution of hepatitis rates. From this analysis using Moranís I and Gearyís c, we must conclude that there is not significant spatial autocorrelation.
Table 2: Output
The input data file: hep.dat
The total number of points: 58 Distance Moran's I Expected I Variance Z-value 50.0000 0.0319 -0.0175 0.0172 0.3776 100.0000 0.0638 -0.0175 0.0095 0.8365 150.0000 0.0704 -0.0175 0.0077 0.9995 200.0000 0.0673 -0.0175 0.0072 0.9980 250.0000 0.0652 -0.0175 0.0070 0.9875 The input data file: hep.dat The total number of points: 58 Distance Geary's c Variance Z-value 50.0000 0.27181 0.700783 -0.86986 100.0000 0.28573 0.455953 -1.05779 150.0000 0.29893 0.391380 -1.12063 200.0000 0.31507 0.365535 -1.13287 250.0000 0.33074 0.354542 -1.12398
References
Cliff, A.D. and Ord, J.K. (1973) Spatial Autocorrelation, Pion: London
Cliff, A.D. and Ord, J.K. (1981) Spatial Processes: Models and Applications, Pion: London
State of California Department of Health Services (March 1999). 1998 Report Health Data Summaries for California Counties